Three-dimensional intact-tissue sequencing of single-cell transcriptional states
Posted on Jul 27, 2018
Abstract
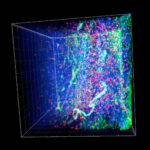
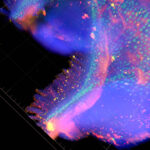
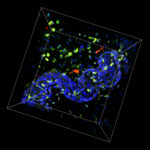
Retrieving high-content gene-expression information while retaining three-dimensional (3D) positional anatomy at cellular resolution has been difficult, limiting integrative understanding of structure and function in complex biological tissues. We developed and applied a technology for 3D intact-tissue RNA sequencing, termed STARmap (spatially-resolved transcript amplicon readout mapping), which integrates hydrogel-tissue chemistry, targeted signal amplification, and in situ sequencing.The capabilities of STARmap were tested by mapping 160 to 1020 genes simultaneously in sections of mouse brain at single-cell resolution with high efficiency, accuracy, and reproducibility. Moving to thick tissue blocks, we observed a molecularly defined gradient distribution of excitatory-neuron subtypes across cubic millimeter–scale volumes (>30,000 cells) and a short-range 3D self-clustering in many inhibitory-neuron subtypes that could be identified and described with 3D STARmap.
By Xiao Wang, William E. Allen, Matthew A. Wright, Emily L. Sylwestrak, Nikolay Samusik, Sam Vesuna, Kathryn Evans, Cindy Liu, Charu Ramakrishnan, Jia Liu, Garry P. Nolan, Felice-Alessio Bava, Karl Deisseroth
Science 27 July 2018, Vol. 361, Issue 6400